Identifying Novel Genes Associated with Autism
Autism is a common childhood neurodevelopmental disorder affecting one in 180 children. It is characterized by impaired social interaction and communication, and by restricted interests and repetitive behavior. Autism is a complex disease exhibiting strong genetic liability with a twenty-five-fold increased risk for individuals having affected first-degree relatives. Moreover, the concordance for developing autism is over 90 percent in identical twins, but only 5–10 percent for fraternal twins. Recent advances in genetics show that autism is associated with many diverse genes, with each gene accounting only for a few percent of cases, as well as complicated multigenic effects.
Researchers at the Simons Center for Systems Biology have been studying autism for the past two years. We have identified novel genes associated with autism. Our approach is to use single nucleotide polymorphism (SNP) genotyping chips that measure differences between individuals and can uncover candidate genes or regulatory elements (which control gene activity) associated with the disease.
Most individuals differ very little from one another across the human genome. SNPs are the largest class of DNA sequence variation among individuals. A SNP occurs when one base out of the four bases used in DNA is exchanged for another base at the same locus, such that the minor allele frequency is at least 1 percent in a given population. SNPs are found at the rate of roughly one out of every 1,000 base pairs of the human genome. These SNPs provide the best chance of detecting genetic variation, both normal and otherwise, between people.
The data set provided by Autism Genetic Resource Exchange is a large collection of such SNP data from over eight hundred families who have two or more autistic children. The pedigree and phenotype of most individuals in the AGRE families, including parents and children, are provided. The genotyping, as assayed on these chips, is compared with similar tests carried out at Children’s Hospital of Philadelphia.
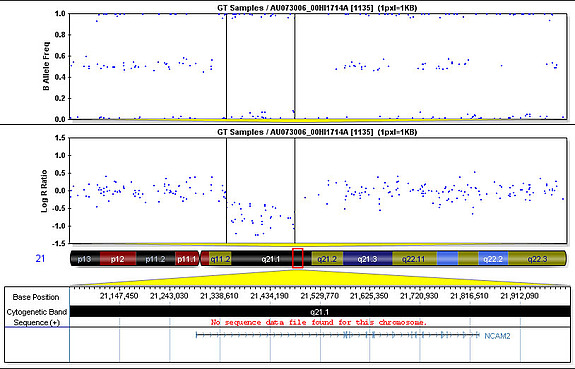
We computed copy number variations (losses or gains of DNA in our genomes, which can result in either a decrease or increase of the number of copies of a gene, partial genes, or multiple genes) and looked for associations with autism. We first identified the copy number variations that have the highest penetrance by looking at autistic and non-autistic siblings in the Autism Genetic Resource Exchange data set. We then tested those copy number variations for an association with autism. We had to collect a large set of data from non-autistic individuals to understand the normal population structure and to test the association.
The controls came from a group of healthy children from the Children’s Hospital of Philadelphia as well as controls used in a study on schizophrenia and type II diabetes. Among the four best candidate genes we found, NRXN1 and CNTNAP2 previously had been found to be associated with autism. Both of these genes produce proteins found in neurons in the central nervous system. The other two genes, NCAM2 and PTPRD, which also encode proteins found in neurons, have not been previously associated with autism. Through experiments in the lab, we have been able to confirm deletions and map their endpoints in DNA samples from children.
This and other studies show that the question “Which genes cause autism?” likely has no simple answer. However, we can ask a different question to probe the mechanism behind this diversity: “Are parents of autistic children more prone to making errors when they copy their DNA to create eggs or sperm?” The errors (such as deletions or duplications of segments of the DNA) are presumably distributed randomly across the genome, exposing the children to all kinds of genetic disorders, including autism. The answer to this question appears to be yes.
The family of genes called p53, p63, and p73 (so named to refer to the relative sizes of the proteins they encode) is involved in ensuring the fidelity of transmission of genetic information when cells replicate their DNA. p53, a tumor suppressor gene, watches over the somatic cells (every cell in our body except eggs and sperm) for DNA damage and errors, protecting us from diseases such as cancer. The genes p63 and p73 protect the genome in eggs produced by females. We find that SNPs in the p63 and p73 genes are over-represented in mothers with autistic children, suggesting that these mothers create a higher rate of copy number variations and other errors in their eggs. This higher rate or frequency of SNPs is not observed in control groups that do not have a family history of autism. Furthermore, the p63 and p73 genes regulate the transcription of other genes that are involved in DNA repair. When the p63 or p73 gene contains a SNP or altered form of DNA, it can produce an altered protein that fails to function properly. This would result in mistakes (poor fidelity) in the DNA of an egg and, in some cases, autism in the offspring.
This is a new approach to studying the genetic causes of autism, because the disease-causing mutations arise spontaneously in the offspring and are not present in the parents or anyone else in the family. The mother or father of autistic offspring may have the SNP that increases the error rate in eggs or sperm that go on to create the affected offspring. And because nothing in this mechanism is special to autism, this concept may play a role in many different genetic disorders. For example, patient populations from in vitro fertility clinics can have problems conceiving due to poor egg quality. We find that the SNPs in p73 that are over-represented in mothers of autistic children are also over-represented in fertility clinic patients. The high error rate associated with this p73 SNP can result in several genetic disorders.
Identifying genes associated with autism gives us a guide to understanding the complex biology of this disorder. The mutations in these genes may someday serve as the basis for diagnostic tests. Ultimately, we hope that genetic studies will provide the basis for a rational and gene-based approach for the treatment or prevention of autism.