The Evolution of New Coronaviruses and the Pandemic
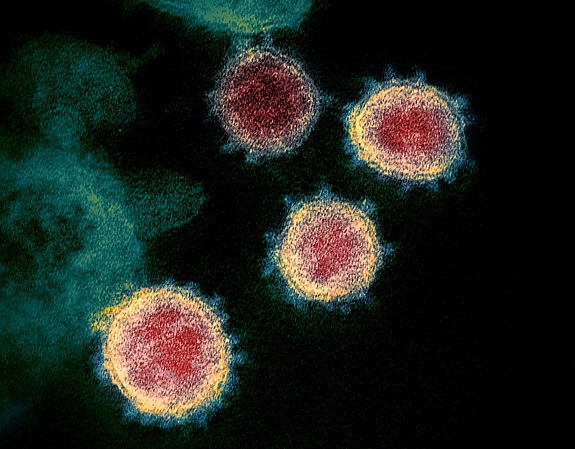
An enormous amount of research on the Covid-19 pandemic is underway. Scientists are trying to understand where the virus is coming from, if related viruses able to create new pandemics are circulating in animal species, how the virus causes disease, and how we can leverage our knowledge to find new therapies, including vaccines. Covid-19 is an infectious disease and, as such, it has to be understood as consisting of two components. The first is the infectious agent, a coronavirus, named SARS-CoV-2. The second component is us, the humans, our human genetics, and our human immune history. In order to understand the disease, we have to understand these two parts and their interaction. Here I will be talking about the virus, what we know about its origin, and how it is evolving.
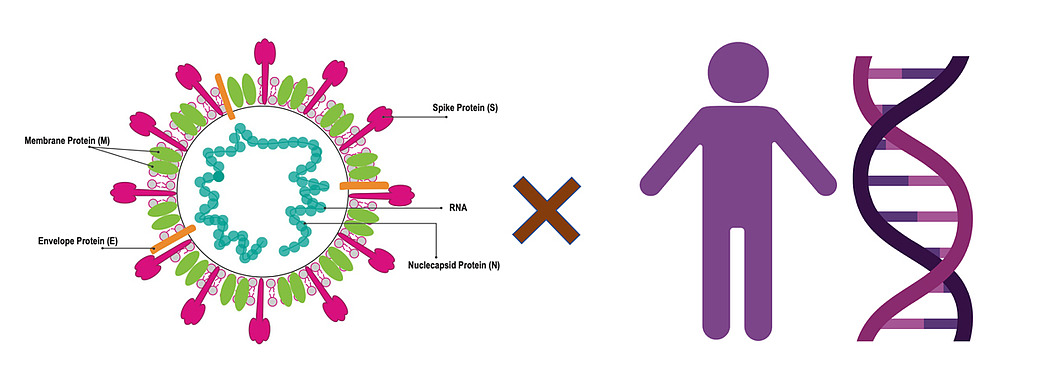
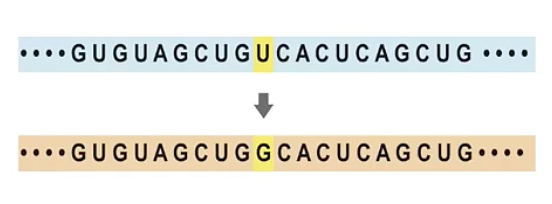
The main theory we have for the emergence of this virus and its evolution is through two mechanisms: mutations and recombination. Mutations refer to mistakes that happen in the viral genome when they are replicating. There is a general rule in life that organisms with very short genomes (like viruses) make many more errors than organisms that have very long genomes (like humans). Mutations in human genomes are very rare because we have a large machinery for correcting errors. Failure to correct these mistakes is one of the common causes of cancer. But viruses accumulate mutations at a much faster pace. The flu, for example, adds one mutation to its genome every week or two. SARS-CoV-2, the virus that causes Covid-19, makes one mistake every two weeks. Mutations accumulate as viruses replicate, creating a diversity of viruses.
SARS-CoV-2 is a member of the vast family of betacoronaviruses, which are found mostly in bats. Bats are frequently found co-infected with a cocktail of different viruses. Groups of researchers have been going around the world collecting samples from bats and sequencing them. The coronaviruses sampled in the last few years provide a background to understand the origins of the new virus.
SARS-CoV-2 genome was sequenced in the beginning of January, 2020. Its entire genome—written as As, Cs, Gs, and Us—is around 30,000 letters long, roughly a ten-page text. Sequencing samples from other persons, bats, and so on produces many small books with ten pages each. To understand the relationship between these genomes we need to organize the information, and a common tool is the phylogenetic tree. Genomes that are very similar—for example, TCGA and TCGC, which have in common a triplet TCG—are placed on the same branch. By gathering many genomes, the information can be organized into a large tree structure. Researchers can plot the evolution of a virus by using mutations to reconstruct who is infecting whom and how the virus is moving and evolving in the population. For instance, there have been many introductions of SARS-CoV-2 to the United States, from Asia, from Europe, that can be traced by following how mutations accumulate during infection and transmission.
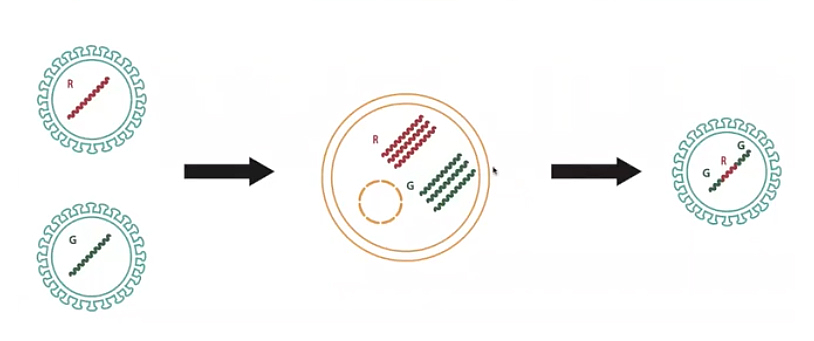
The second mechanism for change in viruses is recombination. If two viruses co-infect the same cell, they produce new genetic combinations called chimeras. Imagine a red virus and a green virus, each with a different genome, that co-infect the same cell. One enters the cell to produce copies of red viral genomes, and the other enters and makes green viral genomes. The chimera produced could be green, red, and green, for instance. This new virus genome is very different from its parents. A child and a parent virus can be very similar in sections of their genome, but differ in some other parts. This recombination process happens pervasively in coronaviruses.
While mutations create small local changes in genomes, recombinations are associated with the acquisition of new genomic material. In general, mixing genomic material at random is rarely successful, but sometimes it enables the virus to acquire new abilities, such as the capacity to infect new hosts. The new coronavirus, SARS-CoV-2, acquired the ability to enter and infect human cells by adapting the spike protein to bind to a particular human protein, ACE2. The same mechanism was used by the SARS-CoV virus in 2002. The viruses responsible for the outbreaks in 2002, and now in 2020, are related in the one particular region that codes for the spike gene that allows the virus to enter into a cell. This potentially indicates that there has been a recombination.
Additional sequencing has found that the closest relative of SARS-CoV-2 is a virus found in horseshoe bats in the south of China in July 2013. What happened from then to now is unknown, but by looking at recombinations and mutations, we can reconstruct the virus and its history. It is like a puzzle: we have many related viral genomes and we need to figure out exactly how those pieces of information came together to generate the pandemic virus.
This virus probably emerged following a two-hit scenario. First, viruses can exchange genetic material through recombination. Recombination enables some viruses to pick up genes that allow them to imperfectly bind to human proteins and infect human cells. We think this happened at least twenty years ago. Since then, there have been further mutations that induce refinements to the ability of the viruses to bind to particular human proteins.
There are many similarities between the SARS outbreak and the current Covid-19 pandemic, and it is very instructive to compare the scientific literature in the years that followed the SARS outbreak to current scientific papers on Covid-19. These two viruses are genetically very, very similar. And the diseases are also similar: immune deregulation, severe respiratory disease, severity increasing with age and more severe in males, etc. We are moving very fast with our investigation into SARS-CoV-2 because of the many things we have learned about the SARS virus.
There are still many missing pieces that we do not understand, but we have a significant amount of information. We now have more than 100,000 SARS-CoV-2 genomes from all corners of the world and we are collecting a significant amount of information on the infected individuals. With the right tools and original ideas, we can elucidate how this virus changes, how it causes disease, and how we can prevent it. Hopefully science will help us to be better prepared in the future.